Many of the methods of molecular biology, including techniques for studying cladistics (patterns of evolutionary relationships) and the molecular clocks used to estimate the time elapsed since two populations diverged, are critically dependent on a hierarchic model of evolution. A genetically closed ancestral population is presumed to have diverged repeatedly, creating a hierarchy of well-defined ancestral lineages (see Figure 2).
Molecular clock methods, for example, assume that mutations accumulate at a more or less steady rate and that lineages that have diverged will never again reconverge. Under these assumptions it is a relatively simple matter to get a rough estimate of the time that has elapsed since two lineages diverged. You just count the differences between the two species' genomes, divide by two and then work out, with reference to the 'background rate' of mutation (how many mutations, on average, per generation), how long it would have taken to reach the current state.
The molecular clock method can only work if the mutations that occur accumulate. In situations where heroic selection pressures are extreme, deleterious mutations will tend to disappear from the genome. The effect of this would be to slow the molecular clock down at times when the system was changing rapidly and speed it up again when the system entered a stable attractor. Natural selection could offset this speeding up and slowing down, of course, but there is a complex co-dynamic feedback to be considered between factors that generate mutations and those that eliminate them.
As if this were not complicated enough, there is now evidence that mutations, far from occurring at random, are clumped in 'hotspots' in the genome and occur at rather different rates in different species. In E. coli, the model organism for much genetic research, scientists have found that across some ~2600 genes neutral mutation rates can vary by a factor of 10 or more (Martincorena et al. 2012). Primates seem to have hotspots and coldspots in their genomes too (Bailey and Eichler 2006) and it is reasonable to conceive of the system in terms of many molecular clocks, each ticking at a different rate. Moreover, if mutations are concentrated at hotspots, it seems reasonable to expect that the same mutation could have happened many times in different populations and, on occasion, could have reversed itself by counter-mutation.
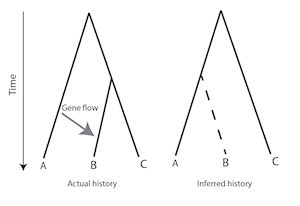
The evidence suggesting reticulated evolution implies that hominin populations were probably vulnerable to periodic crashes, demographic bottlenecks that would have flushed deleterious traits out of hiding and hybridisation events that would move genes between separate lineages. Under these circumstances the molecular clock could speed up, slow down and even run backwards. Figure 4 shows an illustrative example, a tree connecting three species in which lateral gene flow caused by hybridisation drives the molecular clock backwards.
